Overview
Large atomic models (LAM), also known as machine learning interatomic potentials (MLIPs), are considered foundation models that predict atomic interactions across diverse systems using data-driven approaches. LAMBench is a benchmark designed to evaluate the performance of such models. It provides a comprehensive suite of tests and metrics to help developers and researchers understand the accuracy and generalizability of their machine learning models.
Our mission includes:
- Provide a comprehensive benchmark: Covering diverse atomic systems across multiple domains.
- Align with real-world applications: Bridging the gap between model performance on benchmarks and their impact on scientific discovery.
- Enable clear model differentiation: Offering high discriminative power to distinguish between models with varying performance.
- Facilitate continuous improvement: Creating dynamically evolving benchmarks.
LAMBench Leaderboard
Top: Normalized Accuracy of Energy, Force, and Virial Predicting Tasks
Bottom: Accuracy-Efficiency Trade-off
Results are aggregated from all 5 domains of zero-shot prediction tasks. The inference efficiency is displayed as the x-axis of the scatter plot. Other metrics are not visualized here.
Domain Zero-shot Accuracy
We categorize all zero-shot prediction tasks into 5 domains:
- Inorganic Materials: Torres2019Analysis, Batzner2022equivariant, SubAlex_9k, Sours2023Applications, Lopanitsyna2023Modeling_A, Lopanitsyna2023Modeling_B, Dai2024Deep, WBM_25k
- Small Molecules: ANI-1x, Torsionnet500
- Catalysis: Vandermause2022Active, Zhang2019Bridging, Zhang2024Active, Villanueva2024Water
- Reactions: Gasteiger2020Fast, Guan2022Benchmark
- Biomoleculars/Supramoleculars: MD22, AIMD-Chig
To assess model performance across these domains, we use zero-shot inference with energy-bias term adjustments based on test dataset statistics. Performance metrics are aggregated as follows:
-
Metric Normalization: Each test metric is normalized by its dataset's standard deviation:
where:
- is the normalized metric for type on dataset
- is the original metric value (mean absolute error)
- is the standard deviation of the metric on dataset
- denotes the type of metric: E (energy), F (force), V (virial)
- indexes over the datasets in a domain
-
Domain Aggregation: For each domain, we compute the log-average of normalized metrics across tasks:
-
Combined Score: We calculate a weighted domain score (lower is better):
Note: values are displayed on the bar plot of each domain.
-
Cross-Model Normalization: We normalize using negative logarithm:
Note: values are displayed on the radar plot.
-
Overall Performance: The final model score is the arithmetic mean of all domain scores:
Note: values are displayed on the y-axis of the scatter plot.
Efficiency
To evaluate model efficiency, we measure inference speed and success rate across different atomic systems using the following methodology:
Testing Protocol:
- Warmup Phase: Initial 20% of test samples are excluded from timing
- Timed Inference: Measure the execution time for the remaining samples
- Metrics Calculation:
-
Success Rate (): Percentage of completed inferences
-
Time Consumed (): Average time per inference step
-
Efficiency (): Average inference speed in frames/s
-
Benchmark Structure:
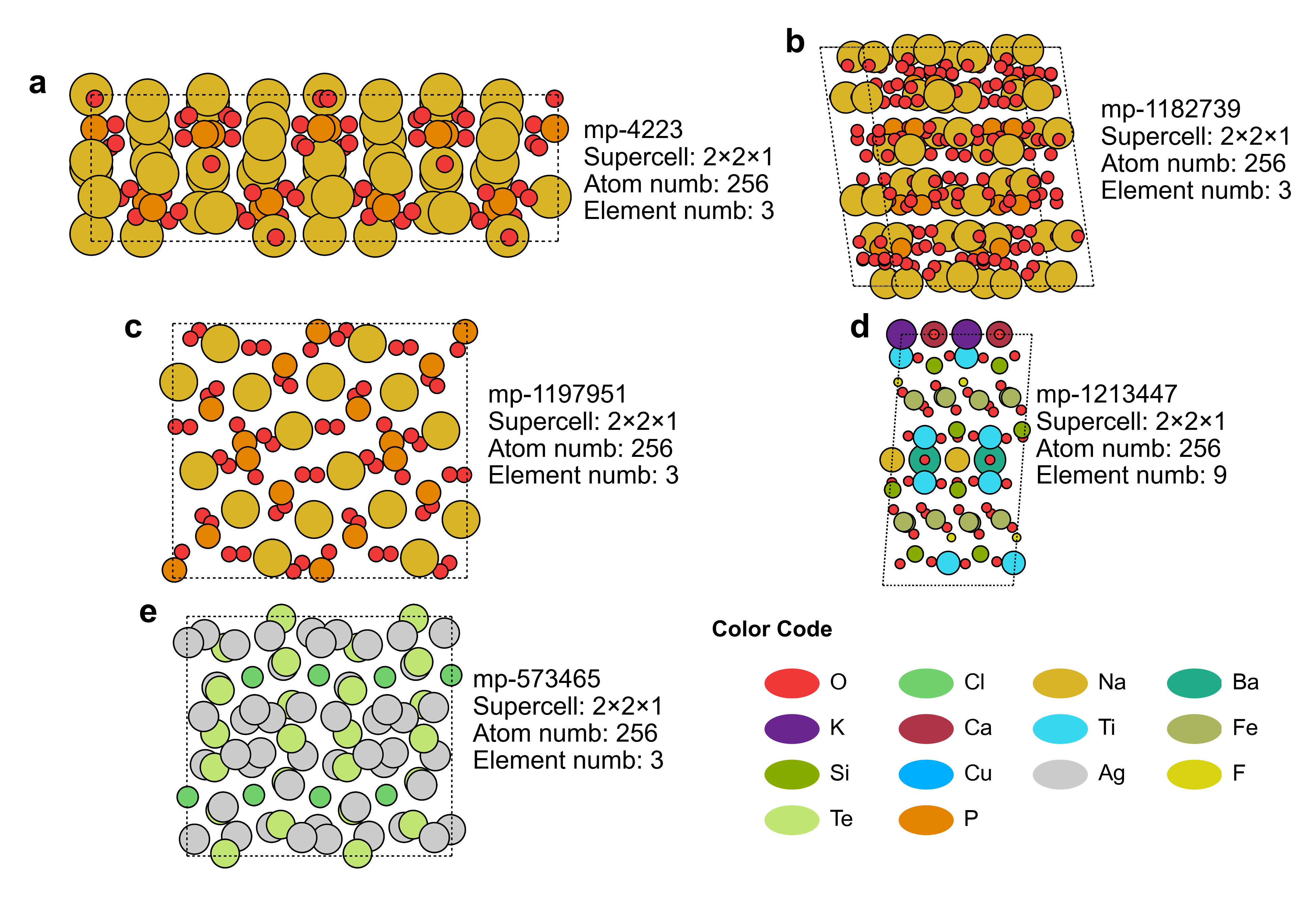
Benchmark structures with five configurations: (a)–(e) feature different elemental compositions while maintaining an identical atom count (N = 256). Each configuration was tested separately, and the average metrics were reported.
